Just a few days remain for OC24 Early Bird Pricing | Don’t Delay!
Your flexible, customizable Clinical Trial Technology Platform
Electronic Data Capture (EDC) & Clinical Data Management
Unrivaled experience for clinicians & patients
- Self-service enablement: Build studies without IT or technical resources
- Smart forms: Create beautiful, ultra-capable eCRFs, featuring real-time edit checks, skip logic and auto-save.
- Simple: Design complex studies on a simple, drag-and-drop interface.
- Data automation: Orchestrate complex data pipelines with ease
EHR eSource Integration
Proven, scalable solution for modern clinical trial research
- Reduce site & clinician burden: Automate outdated processes
- Save time & cost: Reduce downstream SDV
- Early data-drive decisions: Real-time results for sponsors
- Support diverse study types: Clinical trials, RWE, digital health
Reporting Dashboards &
Data Analysis
Actionable, real-time information at your fingertips
- Visualize anything & everything: Out-of-the-box report library
- Custom reports & dashboards: Point & click builder
- Documented tables & fields: Understand and browse data with ease
- Configurable roles: Custom access
Patient Reported Outcomes and Electronic Clinical Outcome Assessments (ePRO and eCOA)
Boost engagement, compliance, data speed & data quality
- Patient-centricity: Remove barriers, simply experience
- Automated notifications: Patients, clinicians & study teams
- Compliance & validation: Validated instruments, ICFs
- Multi-media & multilingual: Access for everyone
Randomization & Supply Chain
Total management right within your eCRFs
- Save time: Integrated into workflow
- Comprehensive & flexible: All types of randomized clinical trials
- Complete sample tracking: Digital monitoring across study lifecycle
- Clear reporting: Sites, PIs and throughout study
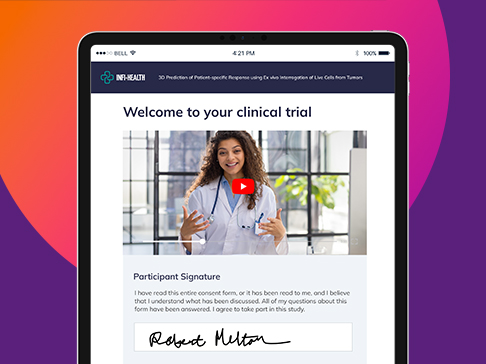
eConsent
Complete eConsent for improved efficiency, patient access & engagement
- Put patients at the center: Remove barriers, deliver expected experience
- Multimedia experience: Support all learning types
- Overcome consent challenges: Multiple trial settings, globalization
- Stop wasting time: Automated workflows support patients & clinicians