We are investing in our customer’s professional growth with the deepest discount possible | Join us IRL at OC24
Stand alone or combine: Whatever works best for your clinical trials.

Electronic Data Capture & Clinical Data Management
- Create beautiful, ultra-capable eCRFs, featuring real-time edit checks, skip logic, and auto-save.
- Design studies on a simple, drag-and-drop interface.
EHR eSource Integration
- Powerfully and easily leverage EHR data for your next study.
- Save countless hours expended by overburdened coordinators and clinicians — reduce routine tasks from hours to seconds!
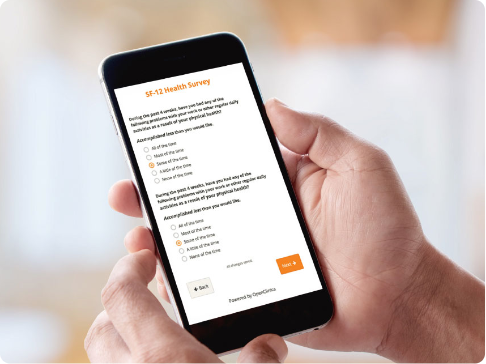
Patient Reported Outcomes and Electronic Clinical Outcome Assessments (ePRO and eCOA)
- Collect clinical patient data faster with user-friendly forms.
- Engage study participants on their own devices regardless of their location.
Randomization & Supply Management
- Add randomization to your studies, with all common randomization methods.
- Track drug kits and monitor inventory in a seamless, integrated user experience.


Reporting Dashboards &
Data Analysis
- Operational and clinical data visualized with bar charts, line graphs, and more.
- Automate the distribution of reports to act quickly to major world events or every day queries.



